run scrublet
Uses Scrublet to predict if an observation (cell) is likely to be a doublet and remove from the dataset. Doublets arise when multiple cells are mistaken as a single cell in droplet-based technologies. This affects biological signal, for example during PCA doublets may form separate clusters not reflecting biological difference.
Parameters
sim_doublet_ratio: float Number of doublets to simulate relative to the number of observed transcriptomes.
expected_doublet_rate: float Where adata_sim not suplied, the estimated doublet rate for the experiment.
stdev_doublet_rate: float Where adata_sim not suplied, uncertainty in the expected doublet rate.
batch_key: str Optional adata.obs column name discriminating between batches.
Web view
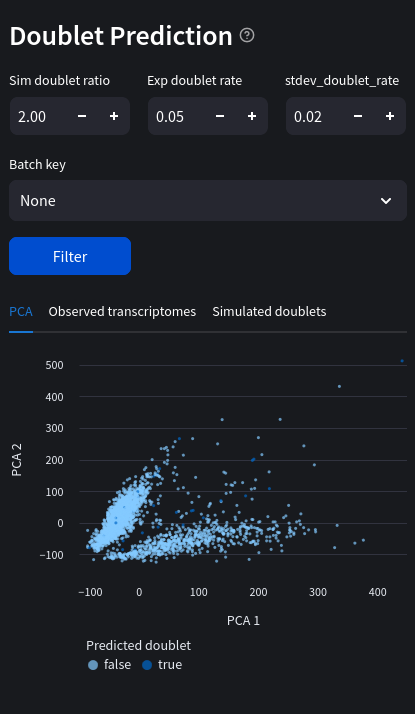
Python equivalent
!pip install scrublet # requires external library scrublet
import scanpy as sc
sc.external.pp.scrublet(adata, sim_doublet_ratio=2, expected_doublet_rate=0.05, stdev_doublet_rate=0.02, batch_key=None, random_state=42)
# plot PCA with doublet predictions
sc.pp.pca(adata)
df = pd.DataFrame({'PCA 1': adata.obsm['X_pca'][:,0], 'PCA 2': adata.obsm['X_pca'][:,1]})
scatter = plt.scatter(x=df['PCA 1'], y=df['PCA 2'], c=adata.obs.predicted_doublet, s=5, label=adata.obs.predicted_doublet.values)
legend1 = plt.legend(*scatter.legend_elements(), loc="upper right", title="Predicted doublet")
plt.ylabel('PCA 2')
plt.xlabel('PCA 1')
plt.title("Predicted doublets in PCA space")
# Simulated doublets and observed transcriptomes prob density plots
sc.external.pl.scrublet_score_distribution(adata)
plt.show()
