batch effect removal
Uses ComBat for batch effect correction [Johnson07]_ [Leek12]_ [Pedersen12]. Corrects for batch effects by fitting linear models, gains statistical power via an EB framework where information is borrowed across genes. This uses the implementation combat.py [Pedersen12]_.
Parameters
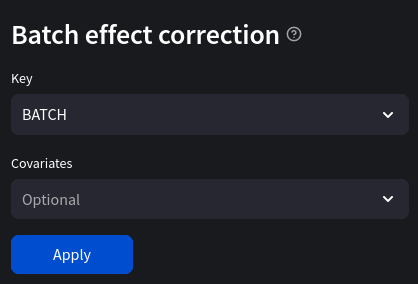
key: str The batch key.
covariates: List[str] Additional covariates besides the batch variable such as adjustment variables or biological condition. This parameter refers to the design matrix X in Equation 2.1 in [Johnson07]_ and to the mod argument in the original combat function in the sva R package. Note that not including covariates may introduce bias or lead to the removal of biological signal in unbalanced designs.
Web view
Python equivalent
import scanpy as sc
sc.pp.combat(adata, key="batch", covariates=None)
