filter highly variable genes
Display genes showing highly variance, optionally remove genes that don't show high variance from the dataset (preserves full genes in raw attribute).
Parameters
flavour: str The method for computing variability in genes. Split between dispersion-based methods [Satija15] and [Zheng17] and normalized variance [Stuart19].
n_top_genes: int Number of top variable genes to display. Mandatory for flavour ‘seurat_v3'.
loess_span: float, default = 0.3 The fraction of the data (cells) used when estimating the variance in the loess model fit if flavor='seurat_v3'.
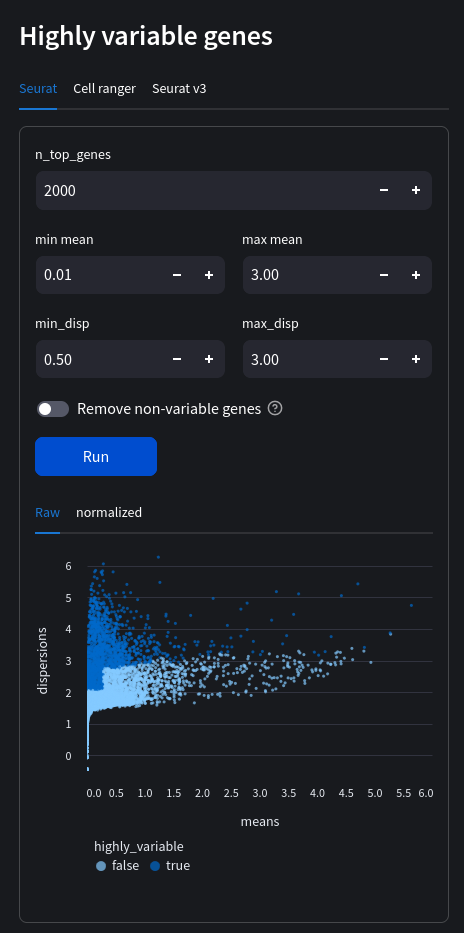
min_mean: float Minimum mean. Ignored if flavour = ‘seurat_v3'.
max_mean: float Maximum mean. Ignored if flavour = ‘seurat_v3'.
min_disp: float Minimum dispersion. Ignored if flavour = ‘seurat_v3'.
max_disp: float Maximum dispersion. Ignored if flavour = ‘seurat_v3'.
remove: bool If set to True, removes non-variable genes from dataset.
Web view
Python equivalent
import scanpy as sc
sc.pp.normalize_total(adata, target_sum=1e4);
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, min_mean=min_mean, max_mean=max_mean, min_disp=0.5)
sc.pl.highly_variable_genes(adata)
